We just posted a new pre-print to bioRxiv on the optimization of Hamiltonian replica exchange simulations. The abstract is below:
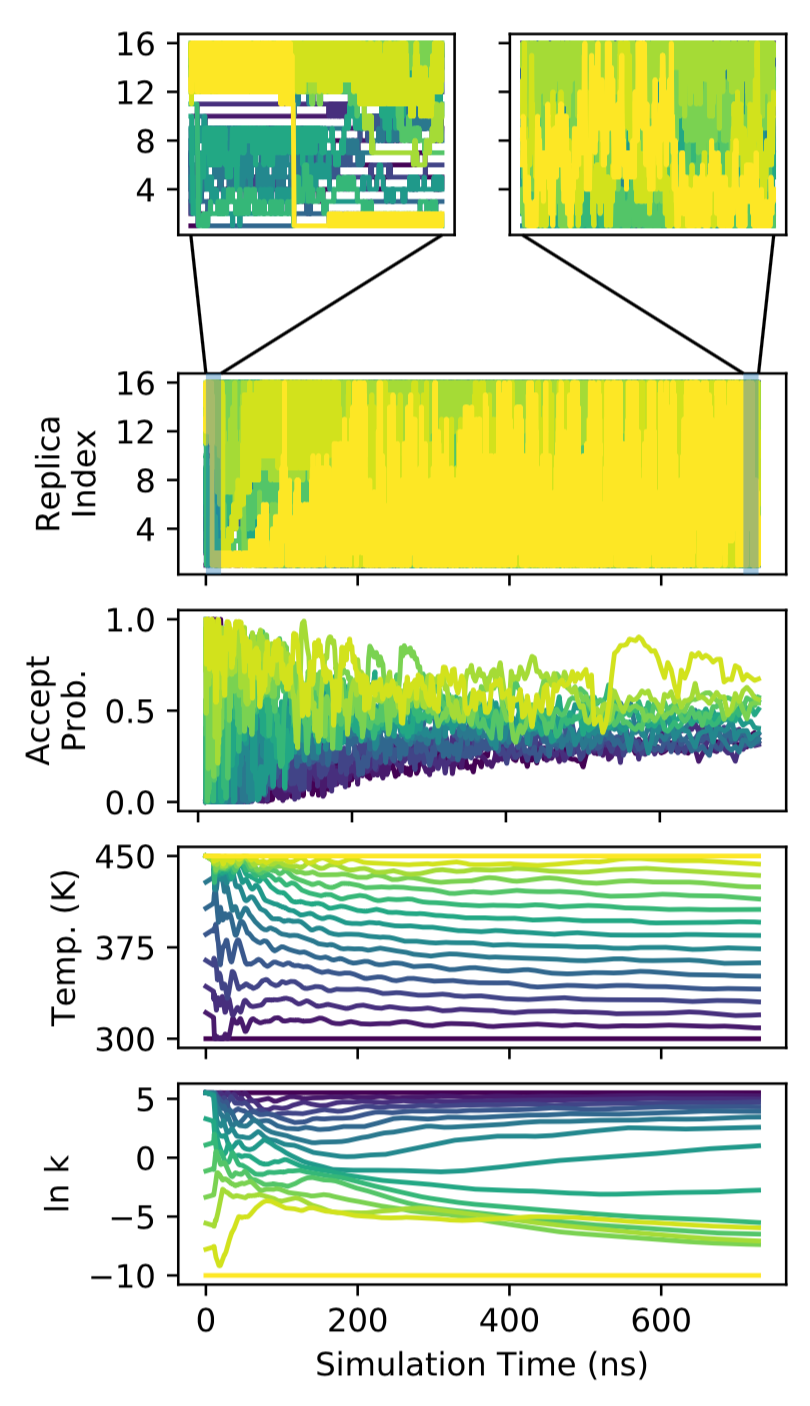
Replica exchange is a widely used sampling strategy in molecular simulation. While a variety of methods exist for optimizing temperature replica exchange, less is known about how to optimize more general Hamiltonian replica exchange simulations. We present an algorithm for the on-line optimization of both temperature and Hamiltonian replica exchange simulations that draws on techniques from the optimization of deep neural networks in machine learning. We optimize a heuristic-based objective function capturing the efficiency of replica exchange. Our approach is general, and has several desirable properties, including: (1) it makes few assumptions about the system of interest; (2) optimization occurs on-line wihout the requirement of pre-simulation; and (3) it readily generalizes to systems where there are multiple control parameters per replica. We explore some general properties of the algorithm on a simple harmonic oscillator system, and demonstrate its effectiveness on a more complex data-guided protein folding simulation.
Feedback is welcome.